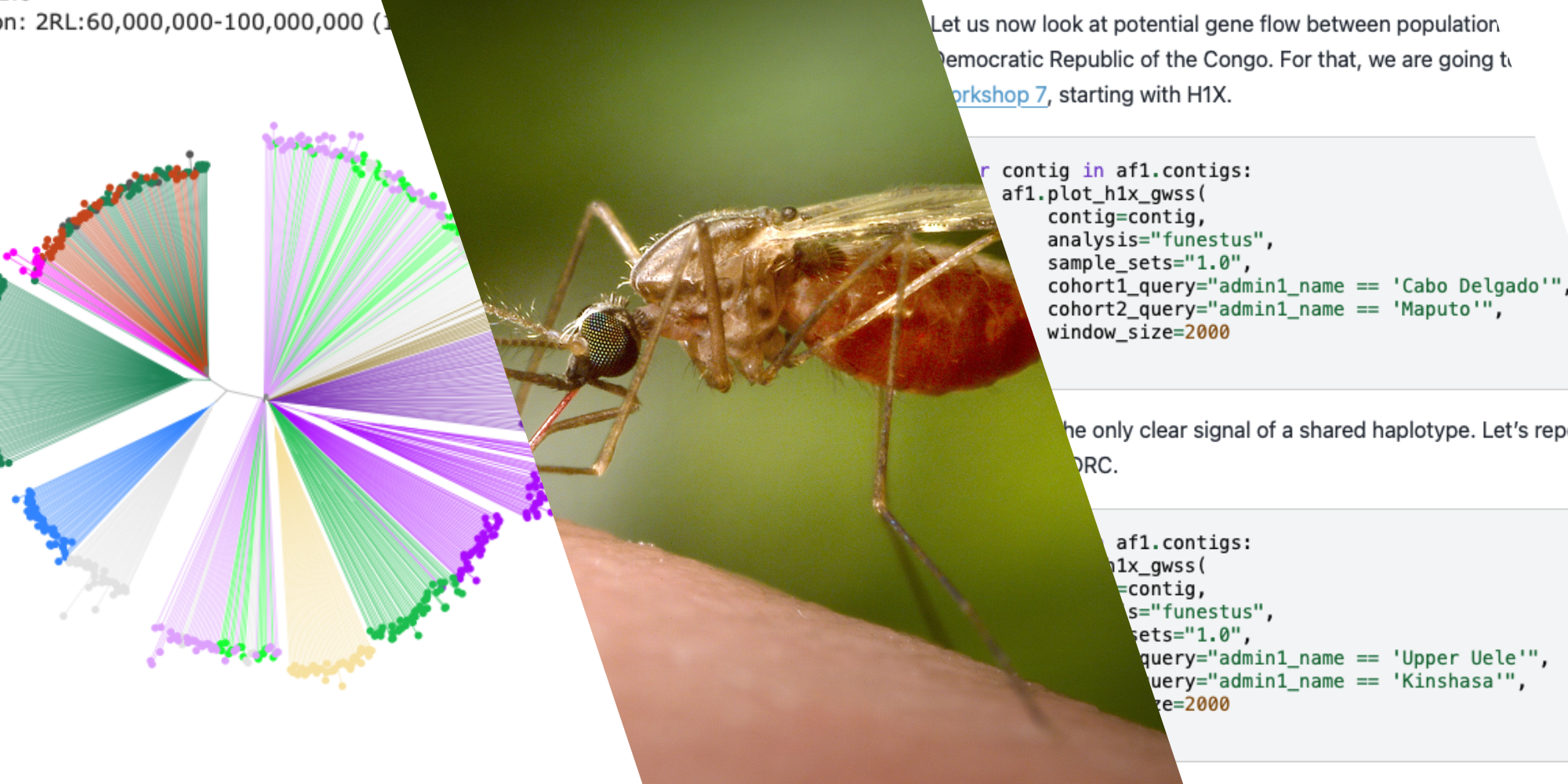
While there are more than 1,500 species of mosquito in the world, only a handful in the Anopheles genus are responsible for malaria transmission. The most famous of these anophelines belong to the Anopheles gambiae complex, including Anopheles gambiae, Anopheles coluzzi, Anopheles arabiensis, and Anopheles quadriannulatus. These are all indistinguishable by eye, and need molecular and genomic techniques to identify precisely.
After decades of research, much about these mosquitoes has been well-characterised. There are several reference genomes with good annotation, many of the genes responsible for insecticide resistance are well-known, and gene drive technology for this species complex has reached an advanced stage.
As a result, most students joining the field of malaria vector genomics start with An. gambiae. Our popular bioinformatics training course is a great example, with 8 workshops that use An. gambiae as an exemplar to introduce essential methods for population genomics.
However, malaria vector patterns are changing.
In some places in Eastern and Southern Africa, it’s Anopheles funestus that is now responsible for the majority of cases of malaria. Unfortunately, the biology of An. funestus is much less well understood. While there is now a reference genome, impressive analysis of 656 whole genomes published in a recent paper in Science, and thousands of samples from the Anopheles funestus genomic surveillance project available through the Malaria Vector Genome Observatory, there are many mysteries to be solved. These include how the mosquito evolves resistance, how its population is structured, and if gene drive technology developed for An. gambiae would work in An. funestus.
To help bridge this gap, the team behind the training course in data analysis for genomic surveillance of African malaria vectors have released three new advanced modules that adapt the tools built to understand the genomic epidemiology of An. gambiae for the An. funestus context.
Not the same mosquito
The first module motivates why separate modules are needed. It explains why studying An. funestus is important, and how it differs from An. gambiae. This comes down to the fact that it’s fundamentally a different species. Compared to An. gambiae, its chromosomes are arranged differently, genes aren’t located in the same place, and the overall level of knowledge about its biology is much lower.
To study An. funestus, you need to leave behind your assumptions built on understanding the genetics of An. gambiae.
“In An. funestus in general, you can assume that the same broad categories of resistance exist, but the genes that are responsible are going to be completely different in almost all cases,” says Jon Brenas, who led on adapting the An. gambiae modules for studying An. funestus.
Detailed analysis
The next two modules explain how to analyse population structure and insecticide resistance genes in An. funestus.
The most common pitfalls, according to Jon, arise from relying too much on familiarity with An. gambiae. “Everything specific that you know about resistance in gambiae is going to be wrong in funestus,” he says.
Those who are familiar with older An. funestus reference genomes may also run into issues locating specific genes or variants. This is because these new modules use the latest reference genome, which is more accurate and complete than its predecessors.
Best of all, these An. funestus modules are integrated into the Malaria Vector Genome Observatory. They are free to use, don’t require downloads, and come with a range of python functions to interrogate the data.
“This resource is easy to use,” says Jon. “All the functions that exist for An. gambiae also exist for An. funestus, with some very small exceptions.”
The An. funestus advanced training modules are available now:
Module 1: Exploring Anopheles funestus
Module 2: Population structure in Anopheles funestus
Module 3: Insecticide resistance in Anopheles funestus
The modules were designed for self-learning and build on concepts from the original course. While they were built to provide useful insights, they are also a work in progress. Please contact the team with issues, questions, comments, and suggestions: support@malariagen.net