These protocols have been developed in partnership with sentinel sites in The Gambia, Ghana, Mali, Vietnam and Indonesia. We are making the laboratory protocols available publicly in case they are useful for other groups.
Before you download any of the protocols, please look at the overview of all the steps in the protocol.
We are currently building capacity to support implementation of protocols by new partners going forwards. In the meantime, if you have questions please contact support@malariagen.net.
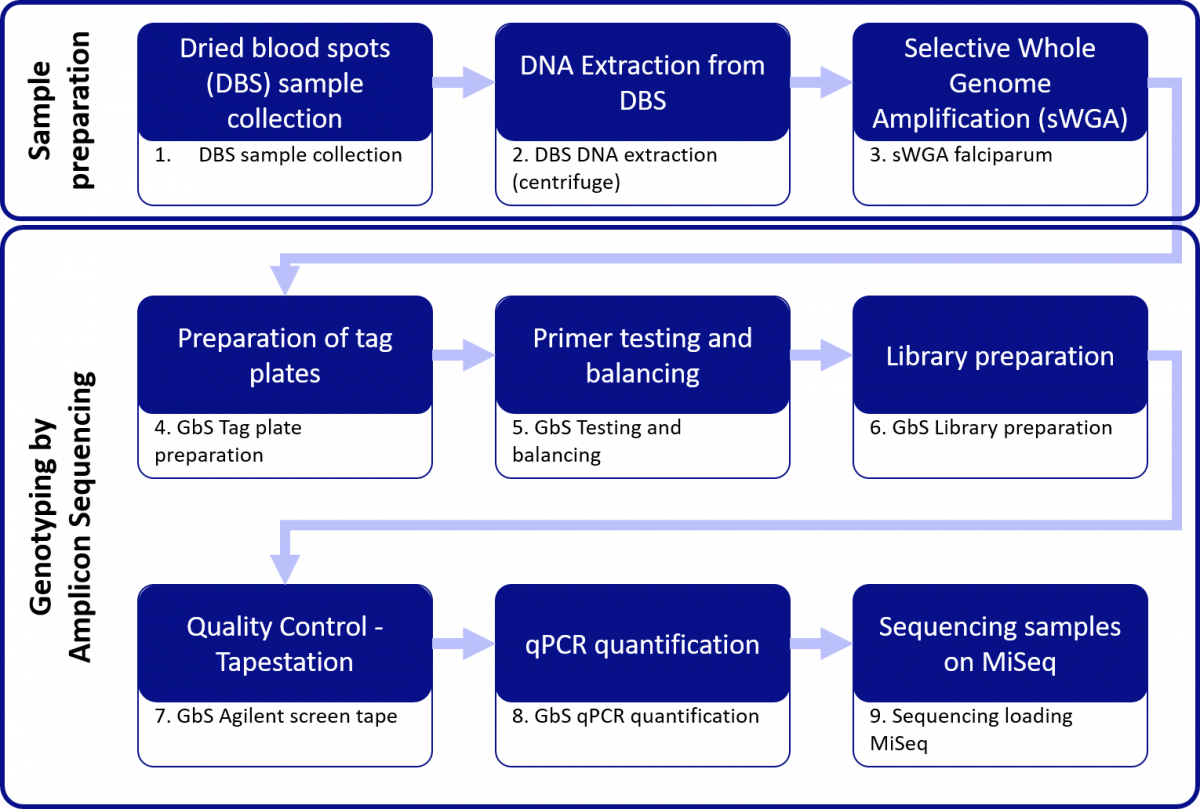
Sample Preparation
- DNA collection – This protocol has been designed to assist our partners collecting dried blood spots from patients, for the purposes of sequencing Plasmodium parasites. Sampling using dried blood spot (DBS) finger-pricks can be performed safely and efficiently and minimises resource and storage requirements compared to sampling with venous blood.
1a. DBS01 DBS Collection Protocol
1b. DBS01 DBS Collection Protocol; Visual Guide
(Please note that the protocols made available for DBS collection were developed as a resource for partners collecting samples to be sent to the Wellcome Sanger Institute for sequencing. These protocols are being modified for in-country sequencing and will be updated.)
- DNA extraction from Dried Blood Spots – This protocol describes isolation of genomic DNA from dried blood spotted onto filter paper following adaptations to a commercially available DNA extraction kit. The procedure yields pure DNA which is suitable for immediate use in amplification reactions.
- DBS02 DBS Extraction (Vacuum Manifold)
- sWGA falciparum – DNA extracted from dried blood spot (DBS) filter papers often has a low parasite DNA yield and an overwhelming human DNA contamination, which poses serious limitations in downstream genetic analyses. Selective whole genome amplification (sWGA) uses short oligonucleotide probes as primers that preferentially bind to the target genome. This protocol describes a method for sWGA that targets Plasmodium falciparum from samples that contain a high proportion of human DNA.
- DBS03 sWGA falciparum
Genotyping by Amplicon Sequencing (GbS)
Please note that these protocols are common across the P. falciparum, P. vivax and mosquito Amplicon Sequencing Toolkit. We are presenting the protocols as an end-to-end process by species, so that in each case there is a full set of protocols.
- Tag plate preparation – This SOP describes the laboratory procedure to produce ‘tag’ plates for use in the PCR_2 step of the GbS process. During the “tagging” process barcode sequences are introduced which allow the clusters derived from different samples to be unambiguously identified on the MiSeq flowcell.
- GbS01 Tag plate preparation
- Testing and rebalancing – These SOPs are used for validation and describe the procedure for determining the optimum concentration of each primer pair in a genotyping by sequencing (GbS) reaction to ensure uniform amplification across the individual targets within the Pf panel.
5a. GbS02a_Testing_and_balancing_Laboratory_protocol_96 AND 5b. GbS02b_Testing_and_balancing_Data_analysis
- Library preparation – This SOP describes the laboratory procedure to capture regions surrounding Single Nucleotide Polymorphisms (SNPs) by high-throughput multiplex PCR, for Illumina MiSeq sequencing. It encompasses the use of validated primer panels, which facilitates even amplification across all targeted loci.
- GbS03 Library preparation (96 well plates) OR 6. GbS03_Library_preparation (384 well plates)
- TapeStation – This is a quality control step which helps establish if the sample purification carried out in the previous step has been successful in “selecting” the correct sized fragments for Illumina MiSeq sequencing.
- GbS04 Agilent ScreenTape
- qPCR quantification – This protocol describes a second quality control step which is used to determine the concentrations of diluted library samples prior to Illumina MiSeq sequencing.
- GbS05 qPCR Quantification
- Illumina MiSeq sequencing– This protocol is an adaptation of the Illumina MiSeq guidelines and describes the method specific to Amplicon Sequencing used to sequence DNA libraries.
- SEQ01 Illumina MiSeq Sequencing
