Malaria causes one in ten of all deaths among children in sub-Saharan Africa, with more than 600,000 dying each year from over 200 million reported cases.
Although the number of deaths is staggeringly high, this is a mortality rate of only 0.3%. Early diagnosis and proper treatment are crucial to prevent death, but it is known that some individuals naturally resist malaria severity.
Why do some infected individuals get very ill, whereas others have no, or very mild, symptoms? Part of our work is to find genetic factors involved and, together with partners in London, Oxford and Tanzania, we have recently discovered three human genetic variants that add to our understanding of the progress of malaria.
We understand only poorly the risk factors for severe malaria, but parasite diversity, host age, immunity and health status are thought to be among those influencing disease progression and outcome.
Variation in human >genes is estimated to account for 25% of this risk. The best example is the beta-globin HBB gene. A particular polymorphism in this gene, known as the HbS allele, is very frequent in Africa. People with two copies of the variant allele suffer from the severe illness sickle cell anaemia; however, people with only one copy of the variant and one normal copy are healthy and less likely to develop severe malaria. But HbS alone cannot explain all the genetic variation involved in the response to the infection. Researchers have found other genes, but these often have weaker effects and, in some cases, different studies find discordant effects for the same variant.
It is clear there is much more to understand and, to uncover other relevant genes, we need detailed and careful assessment of the disease progress in a large group of affected and unaffected children. A team from the Kilimanjaro Christian Medical Centre, Moshi, Tanzania, have recruited 500 children with severe malaria, aged 1–10 years, and compared their genetic variants with a similar group of children who never developed any form of severe malaria.
The team collected detailed descriptions of symptoms, such as severe anaemia, respiratory distress, cerebral malaria and acidosis, in order to discriminate the levels of severity of the malaria episode. The study was conducted in Teule district hospital, Muheza, and surrounding villages, where it is estimated that 165 in 1000 children die before their fifth birthday.
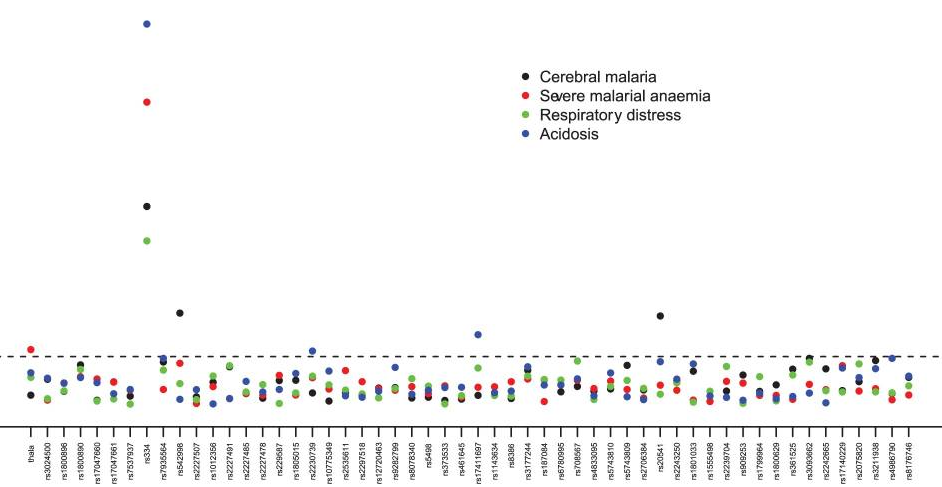
The team looked at the prevalence of 65 human genetic variants (polymorphisms), some reported by other researcher to be associated with altered risk of malaria. This study – genotyping and case-control association analysis – was carried out with the collaboration of teams from the London School of Hygiene and Tropical Medicine, Wellcome Trust Sanger Institute and the Wellcome Trust Centre for Human Genetics, Oxford.
We were able to confirm several known genetic risk factors, including the 90% reduction in severe malaria risk that the HbS allele brings. We also confirmed association between the alpha(+)-thalassaemia polymorphism and severe malaria anaemia, and polymorphisms in two genes on the human X chromosome, G6PD and CD40L, both of which are associated with altered risk of severe malaria in females.
When we looked at the effects on acidosis, we found polymorphisms in the adenylcyclase 9 gene (ADCY9) associated with a decreased risk of acidosis, whereas a polymorphism in the IL1A gene was associated with increased risk.
We found three novel associations between gene variants and malaria severity. Severe malaria was associated with a single base change in an E3 ubiquitin-ligase (TRIM5) gene. Mutations in TRIM5 have been associated with HIV susceptibility, and the finding in malaria is novel.
We showed an increased risk to develop cerebral malaria in people with certain polymorphisms in the reticulon 3 (RTN3) and IL13 genes. Reticulon 3 is a protein made in all cells that is associated with cell membranes and found in a structure called the endoplasmic reticulum. It has been associated with neurodegenerative diseases and is made in lower amounts in brains of mice that are sensitive to Plasmodium berghei (experimental malaria). The IL13 gene is located in a genomic region previously associated with severe malaria (the 5q31–q33 region) in several endemic countries.
Our study confirmed previously reported results and uncovered three new genes. It is important that further studies are performed in Tanzania and in other malaria-endemic regions to test the generality of these associations. Although several genes have been identified that can protect against severe malaria, their functional role still needs to be elucidated. Such follow-up research could lead in the future to improvements in and development of personalised treatments.
Susana Campino is a staff scientist in Dominic Kwiatkowski’s Malaria Genetics Group. Her main interest since her PhD is on malaria genetics in the host and parasite.
Publication
Manjurano et al. Candidate Human Genetic Polymorphisms and Severe Malaria in a Tanzanian Population. PLoS One. 2012;7(10):e47463. doi: 10.1371/journal.pone.0047463. Epub 2012 Oct 29.

