Introduction
SpotMalaria harnesses genomic technologies to monitor the global evolution of malaria parasites, delivering knowledge that will increase the efficiency of malaria elimination and eradication efforts.
Objectives & Coordination
Malaria control and elimination programmes often face practical obstacles that can be tackled more effectively with information about the parasite population.
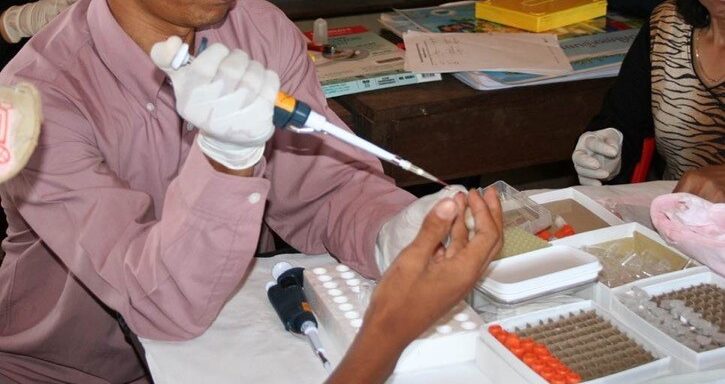
Novel laboratory methods and analytical tools make it possible to rapidly sequence and genotype Plasmodium genomes from dried blood spots (DBS) collected from malaria patients – allowing for more comprehensive coverage in malaria endemic areas.
SpotMalaria connects local and national partners creating a sampling and reporting network for collecting large numbers of dried blood spots and uses a simple standardised operating procedure (SOP) that is easy to implement in the field.
Our main goal is to provide partners with actionable information to support malaria control interventions. We use state-of-the-art genetic and genomic technologies to rapidly genotype Plasmodium falciparum samples isolated from the finger-prick blood samples.
Our SpotMalaria partners receive a Genetic Report Card for each sample that they contribute, showing whether the parasites carry mutations that make them resistant to antimalarial drugs such as artemisinin or its ACT partner drugs. This allows our partners to rapidly detect changes in drug efficacy and to respond to these changes.
Currently, we report on genetic variations that are relevant to resistance to several antimalarial drugs:
- artemisinin (kelch13, arps10, mdr2, fd, crt genes)
- piperaquine (plasmepsin2-3, exo)
- chloroquine, amodiaquine, mefloquine (crt, mdr1)
- sulfadoxine, pyrimethamine (dhfr, dhps)
In addition, we report on other genetic variations relevant to parasite epidemiology:
- Detection of additional species of Plasmodium in a P. falciparum infection, such as P. vivax, P. knowlesi, P. malariae, P. ovale
- Genetic barcodes (101 high-diversity SNPs across the genomes), used to estimate complexity of infection (COI), identify expanding strains and the geographic origins of samples.
We use a set of highly informative single nucleotide polymorphisms (SNPs) to “barcode” parasites, from which we can estimate complexity of infection. We are also able to determine if an infection comprises additional species of Plasmodium, such as P. vivax, P. malariae, P. ovale, and P. knowlesi.
The next project phase will focus on developing easy to use tools that incorporate genetic report card data in the context of the ever-growing database of genotyped malaria parasites. We are also working on methods that allow expansion of genotyping to global partner sites to speed up genotyping time. Additionally, we are working on genotyping methods for typing a similar set of informative loci in P. vivax dried blood spot samples.
Our partners
In the early phases two networks of scientists, clinicians and public health teams have helped refine SpotMalaria. We have expanded our scope and while still working closely with these networks, we now have partners and collaborations from 24 different countries in Asia, Africa, and South America.
GenRe-Mekong
 GenRe-Mekong is funded by the Bill & Melinda Gates Foundation to support malaria elimination efforts in the Greater Mekong Subregion, which straddles regions of Cambodia, Vietnam, Laos, Thailand, and South China. The GenRe-Mekong project aims to supply strategically important information from genetic data to national control programmes and other elimination projects, in the most easily implementable, timely, comprehensive and cost-effective manner possible.
GenRe-Mekong is funded by the Bill & Melinda Gates Foundation to support malaria elimination efforts in the Greater Mekong Subregion, which straddles regions of Cambodia, Vietnam, Laos, Thailand, and South China. The GenRe-Mekong project aims to supply strategically important information from genetic data to national control programmes and other elimination projects, in the most easily implementable, timely, comprehensive and cost-effective manner possible.
Pathogens genomic Diversity Network Africa (PDNA)
The prospect of malaria elimination in Africa presents unique challenges. Malaria is more common and acquired immunity can mean that drug pressure is lower, in addition to the non-biological factors, for example social and political factors, that can affect transmission dynamics. While drug resistance is a large problem, there is also a need to determine how parasite genetics both influence and are influenced by these differences across African populations. To answer these questions, we’re partnering with researchers from the PDNA to establish important baseline genetic data across as many populations as possible.
Ghansah et al. Monitoring parasite diversity for malaria elimination in sub-Saharan Africa. Science, 2014; 345(6202): 1297-8. DOI: 10.1126/science.1259423.
Sampling locations
Bangladesh, Benin, Brazil, Cambodia, Cameroon, Colombia, Congo, Ethiopia, The Gambia, Ghana, Guinea, India, Indonesia, Kenya, Laos, Malaysia, Mali, Myanmar, Peru, Senegal, Sudan, Tanzania, Thailand, Vietnam.
For partners
Resources for partners working on parasites
Acknowledgements
For partners that have publications using data produced by the project we ask that the following statement is included in the acknowledgements.
This publication uses data from the MalariaGEN SpotMalaria project as described in ‘Jacob CG et al.; Genetic surveillance in the Greater Mekong Subregion and South Asia to support malaria control and elimination; eLife 2021;10:e62997 DOI: 10.7554/eLife.62997‘. The project is coordinated by the MalariaGEN Resource Centre with funding from Wellcome (206194, 090770). The authors would like to thank the staff of Wellcome Sanger Institute Sample Management, Genotyping, Sequencing and Informatics teams for their contribution.
Partner studies
Partner study description Effective control and final elimination of malaria in the face of declining transmission and increasing spatial heterogeneity of disease and infection will require the identification and targeting of hotspots or…
Partner study description In The Gambia, malaria transmission has substantially declined in the last 5-10 years and access to treatment is good. The relatively high drug pressure, reduced transmission and consequent waning of…
Partner study description The dry season in The Gambia presents a bottleneck for the total parasite population as there is little or no malaria transmission. Some individuals maintain a malaria infection through this…
Partner study description In terms of malaria prevalence, The Gambia can be divided into two strata: low prevalence in the western part and relatively high prevalence in the eastern part. Nevertheless, within these…
Partner study description A multicentre study to evaluate the in vivo, in vitro and molecular efficacy of artesunate in monotherapy for the uncomplicated malaria treatment is currently running in two study sites in…
Partner study description The study was conducted in localities on the eastern slope of Mt Cameroon, with varying malaria transmission profiles and geographic features. The terrain rises from the Atlantic ocean at the…
Partner study description In the Kassena-Nankana Districts (KNDs) malaria is highly endemic with marked seasonal variation in transmission intensity. The high transmission season coincides with the rainy season, which is from July to…
Partner study description A multi-centre, open-label randomized trial to assess the efficacy, safety and tolerability of Triple Artemisinin-based Combination Therapies (TACTs) compared to Artemisinin-based Combination Therapies (ACTs) in uncomplicated falciparum malaria and to…
Partner study description In terms of malaria prevalence, The Gambia can be divided into two strata: low prevalence in the western part and relatively high prevalence in the eastern part. Nevertheless, within these…
Partner study description This study looks at samples collected from two sites in Ghana. The first is Lekma Hospital, Teshie, Accra; near the coast, an eastern suburb of Accra. Malaria transmission in Teshie…
Partner study description A multi-centre, open-label randomized trial to assess the efficacy, safety and tolerability of Triple Artemisinin-based Combination Therapies (TACTs) compared to Artemisinin-based Combination Therapies (ACTs) in uncomplicated falciparum malaria and to…
Partner study description This study looked at different field sites in Mali representing three different major eco-zones across West Africa that are different with regard to endemicity and current implementation of control strategies:…
Partner study description The objectives of this programme were to scale-up the targeted mass drug administration strategy regionally, and measure its effect on the incidence of clinical malaria in eastern Karen/Kayin state, Myanmar,…
Partner study description This study collected Plasmodium falciparum samples in the Ashanti region of Ghana for genomic sequencing.
1) An ongoing state-wide malaria surveillance study in Sabah, Malaysia funded through NIH and Australian NHMRC. All patients presenting to government health facilities with symptoms of malaria and microscopically confirmed Plasmodium species (with…
Partner study description Genetic surveillance project conducted by the National Malaria Control Programme in partnership with GenRe-Mekong in endemic region of the Lao PDR. This is part of a large project of genetic…
Partner study description Genetic surveillance project conducted by the National Malaria Control Programme in partnership with GenRe-Mekong in endemic region of Vietnam. This is part of a large project of genetic surveillance of…
Partner study description This study aimed to determine risk factors for catching malaria in northeast Thailand, including travel, to identify where people were likely being infected and determine how much antimalarial drug resistance…
Partner study description This study is a country-wide retrospective analysis of molecular markers with potential impact on resistance before and after ACT introduction (18-year period).
Partner study description A prospective study designed to identify molecular markers of antimalarial drug resistance in Plasmodium falciparum in Yola, Nigeria. Objectives of the study are: To detect clinically significant genes associated with…
Partner study description This study and the adjoined Plasmodium vivax study 1250 collectively form a project aimed towards providing genetic and clinical data of circulating P. falciparum and P. vivax populations in different…
Partner Study Description As countries decrease malaria transmission and come closer to elimination, transmission becomes more fragmented and focal, and human population movement becomes an important factor in the spread of infection. Proper…
Partner study description Genetic surveillance project conducted by the National Malaria Control Programme in partnership with GenRe-Mekong in endemic regions of Vietnam. The study aimed to determine risk factors for catching malaria in…
Partner study description Given the emergence of Plasmodium falciparum (Pf) artemisinin resistance Médecins sans Frontières launched in May 2014 a research programme in Chey Saen district (Cambodia), aimed at preventing its spread. The…
Partner study description The main aim of the study shall be to determine the viability and the pathogenicity of Plasmodium falciparum in donor blood units during storage under standard conditions (2-8ᵒC). Specifically, the…
Partner study description This is a cross sectional and screening study to determine the prevalence of drug resistant alleles in Sudanese Plasmodium falciparum field isolates which has been collected from different areas in…
This study and the adjoined Plasmodium falciparum study 1227 collectively form a project aimed towards providing genetic and clinical data of circulating P. falciparum and P. vivax populations in different endemic regions in…
Partner study description Randomized trial to assess the safety, tolerability and efficacy of arterolane-piperaquine, arterolane-piperaquine+mefloquine and artemether-lumefantrine. Number of subjects 219. We obtain parasite clearance half lives and travel surveys.
Partner study description Resistance of Plasmodium species to previous generations of medicines, such as chloroquine and sulfadoxine-pyrimethamine (SP), influenced the replacement of chloroquine (CQ) with sulfadoxine-pyrimethamine (SP) then artemisinin-based combination therapy (ACT) for…
Partner study description The Plasmodium falciparum samples due to be collected from seven countries (The Gambia, Ghana, Mali, Cameroon, Ethiopia, Tanzania and Madagascar) will contribute to the Pan-African Malaria Genetic Epidemiology Network (PAMGEN)…
Partner study description The Plasmodium vivax samples due to be collected from Ethiopia and Madagascar will contribute to the Pan-African Malaria Genetic Epidemiology Network (PAMGEN) project. The aim of PAMGEN is to create…
Malaria remains a major public health problem in Sudan and worldwide. One of the major problems hindering the efforts to combat malaria is the emergence of drug resistant parasites and the subsequent treatment…
Partner study description The main objective of this study is to demonstrate that pregnant women attending Antenatal Care (ANC) clinics in the DRC are a suitable, pragmatic sentinel population for the genetic surveillance…
Partner study description As part of his PhD project, Collins Morang’a collected asymptomatic and symptomatic individuals during the dry and wet season from Navrongo, respectively. Navrongo is a region of high malaria transmission…
Since 2016, the Dominican Republic has implemented large-scale strategies for the elimination of malaria at the national level. To achieve this objective, a precise, rapid and ultrasensitive diagnosis is necessary that, in addition…
Partner study description This proposal has been developed in response to recently published pilot data showing mutations in genes associated with resistance to artemisinins. The overall objective is to characterise genomic variation inPlasmodium…
This study aims to improve the understanding of the seasonal changes in the asymptomatic carriage of P. falciparum, and its links with local malaria transmission dynamics. The study consists in a cohort of…
This study aims to improve the understanding of the seasonal changes in the asymptomatic carriage of P. falciparum, and its links with local malaria transmission dynamics. The study consists in a cohort of…
Despite the gained success in the control, malaria has remained a major public health problem, affecting particularly children aged < 5 years in sub- Saharan Africa. Most of the malaria transmissions occur during…
This study is part of the Genomic Surveillance Hubs in West Africa/NIHR Global Health Research Group project. By better understanding the genomic landscape of malaria parasites and vectors in this region, National Malaria…
In Ghana, malaria is still one of the top five morbidities at various health facilities. Majority of cases of malaria is caused by the parasite, Plasmodium falciparum. A minority of cases are caused…
Publications
- Household clustering and seasonal genetic variation of Plasmodium falciparum at the community-level in The Gambia
Guery et almedRxiv, 2024;- Sustained clinical benefit of malaria chemoprevention with sulfadoxine-pyrimethamine (SP) in pregnant women in a region with high SP resistance markers
Matambisso et alJournal of Infection, 2024; 88 (5)- Molecular surveillance of Plasmodium falciparum drug-resistance markers in Vietnam using multiplex amplicon sequencing (2000–2016)
Rovira-Vallbona et alScientific Reports, 2023; 13 13948- Genome-wide genetic variation and molecular surveillance of drug resistance in Plasmodium falciparum isolates from asymptomatic individuals in Ouélessébougou, Mali
Vanheer et alScientific Reports, 2023; 13(1) 9522- Targeted and whole-genome sequencing reveal a north-south divide in P. falciparum drug resistance markers and genetic structure in Mozambique
da Silva et alNature Communications Biology, 2023; 6 619- Drug resistance and population structure of Plasmodium falciparum and Plasmodium vivax in the Peruvian Amazon
Villena FE et al.Scientific Reports, 2022; 12(1) 16474- Phenotypic characterization of Ghanaian P. falciparum clinical isolates reveals a homogenous parasite population
Thiam et alFrontiers Immunology, 2022; 13 1009252- Genetic surveillance in the Greater Mekong Subregion and South Asia to support malaria control and elimination: about the data
Jacob CG, Thuy-Nhien N, Mayxay M, et al.eLife, 2021; 10:e62997- Temporal evolution of sulfadoxine-pyrimethamine resistance genotypes and genetic diversity in response to a decade of increased interventions against Plasmodium falciparum in northern Ghana
Amenga-Etego, L.N. et alMalaria Journal, 2021; 20(152)- Antimalarial drug resistance molecular makers of Plasmodium falciparum isolates from Sudan during 2015–2017
Hussien et al.PLOS ONE, 2020; 15(8) e0235401- Occurrence of septuple and elevated Pfdhfr-Pfdhps quintuple mutations in a general population threatens the use of sulfadoxine-pyrimethamine for malaria prevention during pregnancy in eastern coast of Tanzania
Bwire et alBMC Infectious Diseases, 2020; 20 530- Efficacy of dihydroartemisinin/piperaquine and artesunate monotherapy for the treatment of uncomplicated Plasmodium falciparum malaria in Central Vietnam
Rovira-Vallbona E et al.Journal of Antimicrobial Chemotherapy, 2020; 75 2272- Mapping the travel patterns of people with malaria in Bangladesh
Sinha et al.BMC Medicine, 2020; 18(1) 45- Detection of mutations associated with artemisinin resistance at k13- propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania
Bwire et alScientific Reports, 2020; 10 3500- Assessment of Plasmodium falciparum drug resistance molecular markers from the Blue Nile State, Southeast Sudan
Mohamed et al.Malaria Journal, 2020; 19(1) 78- Analysis of Plasmodium falciparum Rh2b deletion polymorphism across different transmission areas
Aniweh et alScientific Reports, 2020; 10(1) 1498- Mapping imported malaria in Bangladesh using parasite genetic and human mobility data
Chang et al.eLife, 2019; 8 e43481- Prevalence of chloroquine and antifolate drug resistance alleles in Plasmodium falciparum clinical isolates from three areas in Ghana
Abugri et alAAS Open Research, 2018; 1 1- Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification
Oyola et. al.Malaria Journal, 2016; 15 597
Project contact